|
Based on the PIC values,
it was found that 93% of these markers showed values of more than 0.5, indicating
that these microsatellite markers can be effectively used for molecular characterization
and genetic diversity studies. The results of the X2 test of goodness of fit revealed that
the population was in Hardy-Weinberg Equilibrium (HWE) proportions for 19
microsatellite loci. The remaining eight loci departed from HWE. The observed heterozygosity
ranged from 0.6250-0.8462 with a mean of 0.7404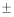 0.06 while the expected
heterozygosity ranged from 0.7211-0.8422 with a mean of
0.8106 0.06 while the expected
heterozygosity ranged from 0.7211-0.8422 with a mean of
0.8106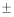 0.03. Population inbreeding estimate
(FIS) indicates heterozygote deficiency was observed to be 0.0666 with a range from
- 0.0063 (OarHH72)-0.2233 (OarJMP29). Though positive
FIS values were observed in
20 loci, they reflect inbreeding in Coimbatore sheep. The markers used in the study
were highly informative and high heterozygosity value is indicative of the higher amount
of genetic variability that can be exploited for their improvement. 0.03. Population inbreeding estimate
(FIS) indicates heterozygote deficiency was observed to be 0.0666 with a range from
- 0.0063 (OarHH72)-0.2233 (OarJMP29). Though positive
FIS values were observed in
20 loci, they reflect inbreeding in Coimbatore sheep. The markers used in the study
were highly informative and high heterozygosity value is indicative of the higher amount
of genetic variability that can be exploited for their improvement.
In India, there are 42 recognized breeds of sheep distributed in different
agroclimatic zones of the country. Tamil Nadu state is endowed with eight breeds of sheep,
of which five are hairy meat type viz., Madras Red, Mecheri, Kilakarsal, Ramnad
White and Vembur and three are wool type viz., Coimbatore, Trichy Black and Nilagiri
(Acharya, 1982).
Coimbatore sheep, also known by the synonyms, Kurumbai Adu and Coimbatore
Kurumbai, produce coarse carpet wool in addition to their use mainly as meat animals. The
breeding tract of Coimbatore sheep is Coimbatore district of Tamilnadu, flocks are highly
migratory and medium-sized animals with compact body are penned for manure. They are white
in color with varying extent of black or tan coat and fleece in the head and neck, which
may also extend up to the shoulder or back (Figure 1). They are docile and hardy with
faster growth rate and early maturity.
The first step towards conservation of livestock genetic resources is the
genetic characterization with respect to phenotypic parameters, unique qualities and
utility. Subsequently, finding out the genetic architecture through molecular means
and evolutionary relationship with other related breeds would provide valuable
information about the breed for taking up conservation measures. Considering these facts, the
present study was carried out to characterize the Coimbatore breed of sheep using the
molecular marker, such as microsatellites. |





